I have recently written, for WIREs Computational Molecular Science, a review article on the use of Principal Component Analysis (PCA) in the study of dynamical systems, with a particular focus on molecular dynamics (MD) simulations of biomolecules [1]. The aim of this work is to provide a clear and practical overview of how PCA has become a central tool for extracting meaningful collective motions from high-dimensional simulation data, and how modern methodological extensions continue to expand its capabilities.
Continue readingPublications
A Virtual Microscope for Nanoscience
I am pleased to announce the publication of the second edition of my book chapter: “A Short Introduction to the Molecular Dynamics Simulation of Nanomaterials” [1] in Micro and Nanomanufacturing, Volume II, edited by W. Ahmed and M. J. Jackson, Springer, 2025. This new edition reflects both the rapid evolution of molecular dynamics (MD) simulations over the past decade and their growing role in nanoscience.
Molecular dynamics simulations have become a cornerstone of modern nanoscience. They allow us to observe matter at the atomic scale, following the motion of thousands—or millions—of atoms in time, effectively turning the computer into a virtual microscope. From nanoparticles and nanotubes to polymers, membranes, and bio–nano interfaces, MD simulations provide insights that are often inaccessible to experiments alone. They help us understand:
- Structural organization at the nanoscale
- Dynamic processes such as adsorption, diffusion, and self-assembly
- Thermodynamic and mechanical properties relevant to material design
This chapter is written with the explicit goal of making these ideas accessible, without sacrificing physical rigor.
Continue readingHow Surfactant Chain Length Shapes Protein Binding
Surfactants are everywhere in protein science — from biochemical laboratories to industrial detergents. Among them, sodium dodecyl sulfate (SDS) is perhaps the most famous (or infamous), widely used for its ability to bind, deactivate, and often denature proteins. Despite decades of experimental and theoretical work, the molecular details of how surfactants bind to protein surfaces are still not fully understood. In my recent study, “Binding Dynamics of Linear Alkyl-sulfates of Different Chain Lengths on a Protein Surface” [1], I have explored this problem using molecular dynamics (MD) simulations, focusing on how the length of the surfactant’s hydrocarbon chain influences protein binding.
Continue readingChlorophyll in Tight Spaces: How Silica Nanoconfinement Stabilizes Photosynthetic Pigments
In a recent molecular dynamics study [1] in collaboration with Prof. K. J. Karki (Department of Physics, Guangdong Technion-Israel Institute of Technology in China), we explored how EthylChlorophyllide a behaves when confined between two silica surfaces — a situation relevant for artificial photosynthesis, nanomaterials, and bio-inspired light-harvesting systems. Chlorophylls are among the most important molecules on Earth. They enable plants, algae, and photosynthetic bacteria to convert sunlight into chemical energy. Yet, outside their natural protein environment, chlorophyll molecules are fragile as they can easily lose their central magnesium ion (demetallation), they degrade under light, and they tend to aggregate uncontrollably in solution.
In natural photosynthetic systems, proteins protect and organize chlorophylls. Reproducing this level of control in artificial systems remains a major challenge. One promising strategy is nanoconfinement — trapping chlorophyll derivatives inside well-defined inorganic structures such as silica nanopores.
Continue readingLook at the Rainbow in a Soap Film: A simple STEM Project
My heart leaps up when I behold
A rainbow in the sky:
So was it when my life began;
So is it now I am a man;
So be it when I shall grow old,
Or let me die!
The Child is father of the Man;
And I could wish my days to be
Bound each to each by natural piety.William Wordsworth, March 26, 1802
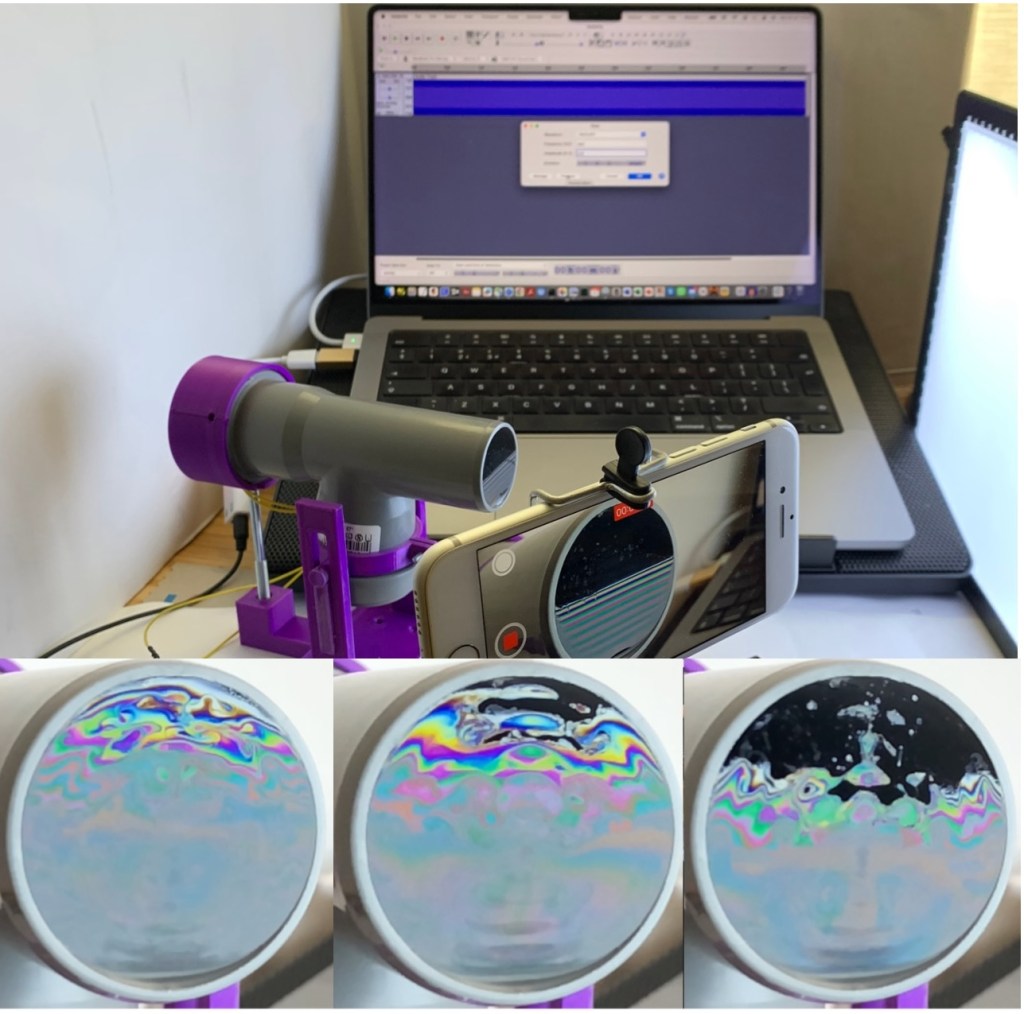
I couldn’t resist citing the beautiful poetry by Wordsworth about the rainbow to introduce my new Instructable, ‘Explore the Physics of Soap Films with the SoapFilmScope.’ I got the idea for this project by reading an article by Gaulon et al. [1]. The authors describe in detail the use of soap film as an educational aid to explore interesting effects in the fluid dynamics of this system. In particular, they examine the impact of acoustic waves on the unique optical properties of the film. In this Instructable, we have designed a device called the SoapFilmScope to perform these experiments. This tutorial will guide you through the process of creating this device, showcasing the mesmerizing interaction between sound waves and liquid membranes. The SoapFilmScope offers an engaging way to explore the physics of acoustics, light interference, and fluid dynamics.
When a sound wave travels through the tube and vibrates the soap film, it creates dynamic patterns through several fascinating mechanisms:
The device consists of a vertical soap film delicately suspended at the end of a tube obtained from a PVC T-shaped fitting that you can get from any DIY store. By attaching a small inexpensive speaker to it, you can let the film dance to the rhythm of the music.
Exploring Disordered Proteins: A Study on Flexible Peptides
Proteins are not always rigid structures. Many of their most important parts — linkers, loops, and disordered regions — are highly flexible, constantly changing shape in solution. To understand how these regions behave, scientists often study short model peptides that capture the essential physics of flexibility. In a new article [1], I have explored the behavior of glycine- and serine-rich octapeptides using molecular dynamics simulations combined with concepts from FRET (Förster Resonance Energy Transfer) spectroscopy (see also my previous post).
Continue readingSeminar Series: Molecular Dynamics Simulation of Biomolecules
In this new series, I will post slides of seminars or lessons that I have delivered in the past years. Some of the reported information is updated, but still helpful. In some cases, I have added descriptions of the slide contents or references to other articles or the original paper where I describe my research results.
I hope you like the presentation, and remember to add your feedback and subscribe to have email notifications about my new blog posts.
In 1648, Isaac Newton published his first edition of the Principia Mathematica, one of the greatest scientific masterpieces of all time. On page 12 of this magnum opus, the famous three laws that bear his name and from which classical mathematical physics evolved are enunciated. More than 350 years after that publication, the same laws formulated to explain the motion of stars and planets remain valuable for us when trying to simplify the description of the atomic world. In the first decades of the last century, the birth of quantum mechanics marked the beginning of the detailed description of atomic physics. The equation of Schrödinger, to the same extent as Newton’s equations, allowed for the mathematically elegant formulation of the shining theoretical intuitions and the experimental data accumulated in the previous decades. Although this equation could be used in principle to describe any molecular system’s physicochemical behaviour, it is impossible to resolve analytically when the number of electrons is more than two. The invention of electronic computers after World War II facilitated the numerical solution of this equation for polyatomic systems. However, despite the continuous and rapid development of computer performance, the ab-initio quantum-mechanical approach to describe static and dynamic properties of molecules containing hundreds or even thousands of atoms, as for biological macromolecules, is still far from becoming a standard computational tool. This approach requires many calculations that can be proportional to , where N is the total number of electrons in the system. It was clear that a reduction, using ad hoc approximations, of the description of the dynamic behaviour of atoms using a classic physics model would be necessary to overcome this problem. In the classical representation, the electrons on the atoms are not explicitly considered, but their mean-field effect is taken into account. Alder and Wainwright performed the first simulation of an atomic fluid using this approximation approximately 63 years ago (1957). They developed and used the method to study simple fluids by means of a model representing atoms as discs and rigid spheres. These first pioneer studies mark the birth of the classical molecular dynamics (MD) simulation technique. The successive use of more realistic interaction potentials has allowed obtaining simulations comparable to experimental data, showing that MD can be a valuable tool for surveying the microscopical properties of physical systems. The first simulations of this type were carried out by Rahman and Verlet (1964): in these simulations, a Lennard-Jones-type potential was used to describe the atomic interactions of argon in the liquid state. Another significant hallmark in this field was the simulation of the first protein (the bovine pancreatic trypsin inhibitor) by McCammon and Karplus in 1977. In the following years, the success obtained in reproducing structural properties of proteins and other macromolecules led to a great spread of the MD within structural biology studies. The continuous increase of computer power and improvement of programming languages has concurred with further refinement of the technique. Its application was progressively expanded to more complex biological systems comprising large protein complexes in a membrane environment. In this way, MD is becoming a powerful and flexible tool with applications in disparate fields, from structural biology to material science.
Nanoparticles in Biology and Medicine
I am very pleased to announce that the second edition of the book Nanoparticles in Biology and Medicine edited by Enrico Ferrari, Mikhail Soloviev is now out.

This fully updated volume presents a wide range of methods for synthesis, surface modification, characterization and application of nano-sized materials (nanoparticles) in the life science and medical fields, with a focus on drug delivery and diagnostics. Beginning with a section on the synthesis of nanoparticles and their applications, the book continues with detailed chapters on nanoparticle derivatization, bio-interface, and nanotoxicity, as well as nanoparticle characterization and advanced methods development. Written for the highly successful Methods in Molecular Biology series, chapters include introductions to their respective topics, lists of the necessary materials and reagents, step-by-step, readily reproducible laboratory protocols, and tips on troubleshooting and avoiding known pitfalls. Authoritative and cutting-edge, Nanoparticles in Biology and Medicine: Methods and Protocols, Second Edition serves as an ideal guide for scientists at all levels of expertise to a wide range of biomedical and pharmaceutical applications including functional protein studies, drug delivery, immunochemistry, imaging, and more.
I have contributed with a chapter (14) titled The Molecular Dynamics Simulation of Peptides on Gold Nanosurfaces.
In this chapter a short tutorial on the preparation of molecular dynamics (MD) simulations for a peptide in solution at the interface of an uncoated gold nanosurface is given. Specifically, the step-by-step procedure will give guidance to set up the simulation of a 16 amino acid long antimicrobial peptide on a gold layer using the program Gromacs for Molecular Dynamics simulations.
Platonic Solid and Chemistry: the Icosahedral Boron Clusters
Boron is the fifth element in the periodic table. It is also the first element of the boron group or the group thirteenth of the periodic table. It is a metalloid element, meaning its properties are between a metal and a nonmetal. The chemical symbol for boron is B, which has an atomic weight of 10.81 grams per mole.

In its ground state, a boron atom contains five electrons arranged in two energy levels. The first energy level, or shell, can hold up to two electrons, while the second can hold up to eight. However, boron has three valence electrons, meaning only the first two energy levels are filled, leaving the third energy level partially empty.
Continue readingLa simulazione di Dinamica Molecolare
- Lex. I. Corpus omne perseverare in statu suo quiescendi vel movendi uniformiter in directum, nili quatenus a viribus impressis cogitur statum illum mutare.
- Lex. II. Muationem motus proportionalem esse vi motrici impressae, et fieri fecundum lineam rectam qua vis illa imprimitur.
- Lex. III. Actioni contrariam semper et equalem esse reactionem: sive corporum duorum actiones in se mutuo semper esse aequales et in partes contrarias dirigi.
Isaac Newton. Philosophiae Naturalis Principia Mathematica. London, 1686.
